Research
Microbiomes are fascinatingly complex – from stochasticity between single cells to disorder among hundreds of interacting species. Our lab searches for quantitative principles emerging from this complexity by integrating theory and high-throughput experiments. Key theoretical approaches include mathematical models of eco-evo dynamics and their analysis using tools from statistical physics. Experimental approaches include in vitro assembly of gut microbiotas and their analysis using omics. Ongoing research projects include:
Coarse-graining microbiomes
Simple quantitative principles can emerge from the complexity of microbiomes.
- Selection can increase functional similarity across microbiomes (Ho and Huang, Cell Syst 2025). We are solving the inverse problem of identifying selected functions from metagenomes.
- Microbial metabolic networks can be coarse-grained both in the lab and in theory. More to come soon!
Quantifying microbial ecology
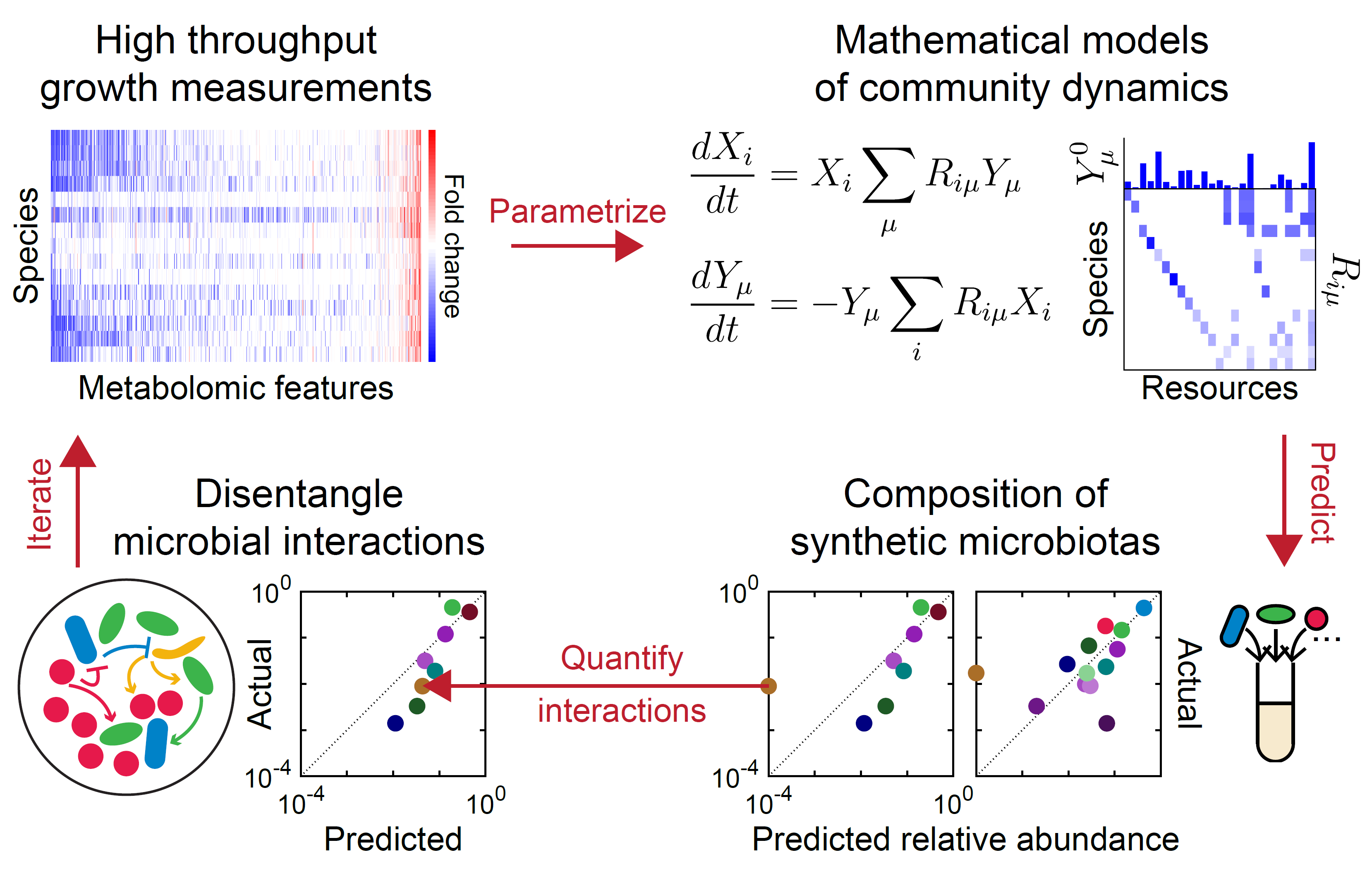
Microbes interact via a plethora of mechanisms, including resource competition, metabolic cross-feeding, and environmental modulation. We disentangle and quantify this complexity using community assembly experiments, omics, and mathematical models.
- The complex dynamics of microbiomes can be predicted by coarse-grained models of resource competition (Ho et al, Nat Microbiol 2024). We are applying this approach to investigate microbial interactions in complex environments and during evolution.
- Microbiomes exhibit special statistics of fluctuations in species abundances, and these fluctuations can be reproduced by typical ensembles of resource competition (Ho et al, eLife 2022). We are using this link to infer the ecological parameters of various ecosystems.
Engineering the human gut microbiome
To quantitatively engineer microbiome composition or function, we build theoretical and experimental tools with focus on the human gut microbiota.
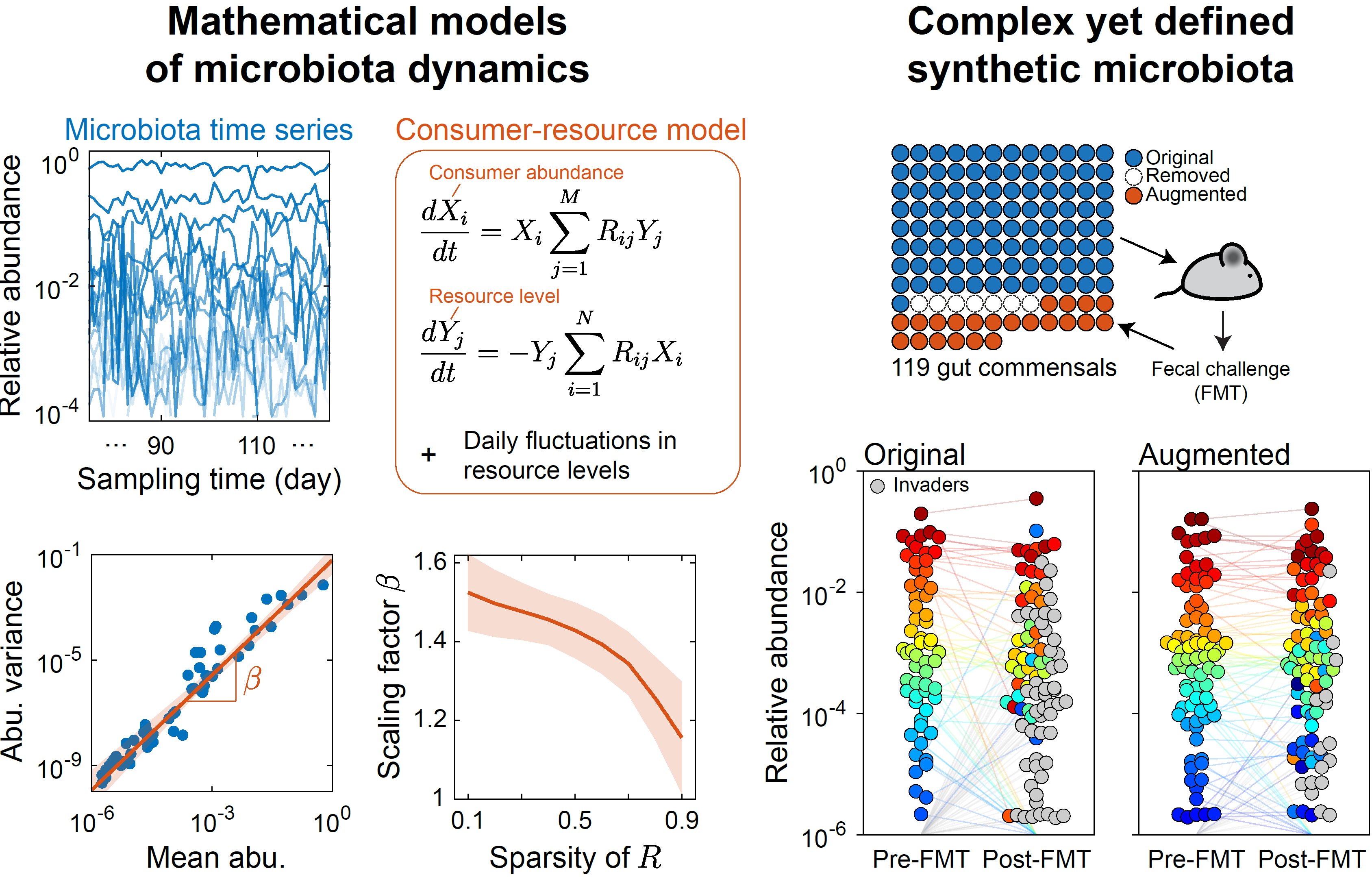
- A highly diverse yet defined model community reproducibly colonizes mice gut, and mice colonized by either the model community or human fecal communities are phenotypically similar (Cheng et al, Cell 2022). We are quantifying microbial interactions in this model system.
- Antibiotic activity can affect the resource competition landscape and vice versa, and this interplay can result in synergism or antagonism among multiple antibiotics (Newton et al. Nat Commun 2023). We are leveraging these predictions to engineer community dynamics.
Microbial cell cycle regulation

How cells regulate and coordinate their growth and cell cycle to maintain a homeostatic cell size is a fundamental question. We use microfluidic-enabled imaging to capture how single cells grow and divide in face of stochasticity, and analyze the data by constructing mathematical models of gene regulatory networks to understand the design principles underlying microbial cell cycle regulation.
- A simple quantitative law describes cell size control in microbes from all domains of life (Ho et al, Annu Rev Biophys 2018). How and why this striking generality arises remain outstanding open questions.
- Cyanobacteria also follows the general law identified above, except for a modification by the circadian clock (Ho et al, Biophys J 2020). How does this coupling affect population fitness?